-
 Github -
MIDST-Lab's Github page.
Github -
MIDST-Lab's Github page.
-
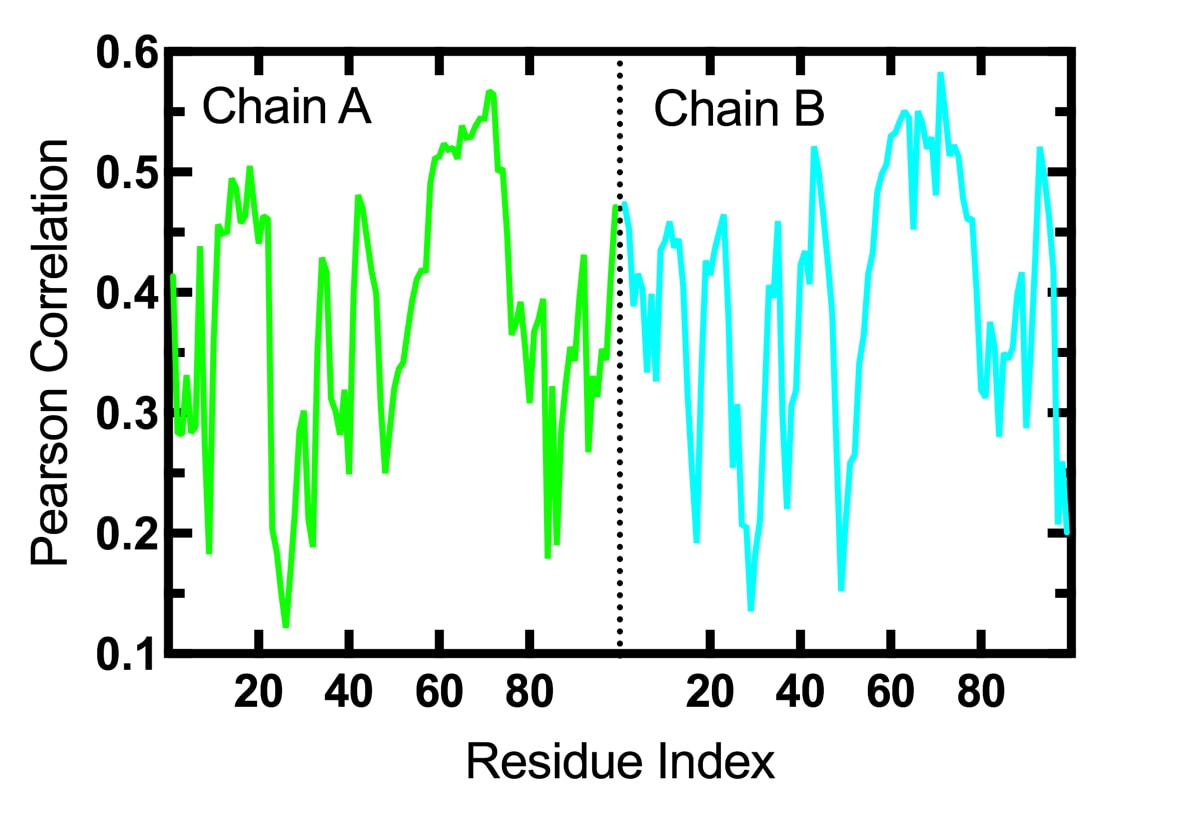 MD-TASK - Suite of Python
scripts that have been developed to analyze molecular dynamics trajectories.
MD-TASK - Suite of Python
scripts that have been developed to analyze molecular dynamics trajectories.
Developed Python scipts fall into 3 categories:
- Residue Interaction Network (RIN) analysis.
- Perturmabion Response Scanning
- Dynamic Cross-Correlation
-
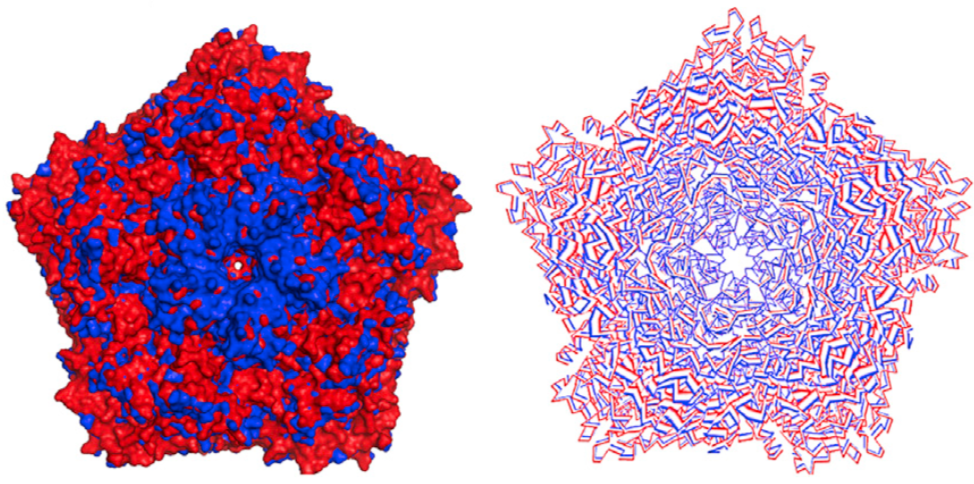 MODE-TASK - Collection of
tools for analysing normal modes and performing principal component analysis on
biological assemblies.
MODE-TASK - Collection of
tools for analysing normal modes and performing principal component analysis on
biological assemblies.
Novel coarse graining techniques allow for analysis of very large assemblies without the need for high performance computing clusters and workstations.
PyMol Plugin
A PyMol Plugin for MODE-TASK has been made available here
-
 Machines - List of machines and servers
used in the MIDST computational laboratory.
Machines - List of machines and servers
used in the MIDST computational laboratory.